Better Confidence Intervals for Quantiles
\[ \newcommand{\bm}[1]{\boldsymbol{\mathbf{#1}}} \DeclareMathOperator*{\argmin}{arg\,min} \DeclareMathOperator*{\argmax}{arg\,max} \]
Abstract
We discuss the computation of confidence intervals for the median or
any other quantile in R. In particular we are interested in the
interpolated order statistic approach suggested by Hettmansperger
and Sheather (1986) and Nyblom (1992). In order to make the
methods available to a greater audience we provide an implementation of
these methods in the R package quantileCI and a small
simulation study is conducted to show that these intervals indeed have a
very good coverage. The study also shows that these intervals perform
better than the currently available approaches in R. We therefore
propose that these intervals should be used more in the future!
 This work is licensed under a Creative Commons
Attribution-ShareAlike 4.0 International License. The
markdown+Rknitr source code of this blog is available under a GNU General Public
License (GPL v3) license from github.
This work is licensed under a Creative Commons
Attribution-ShareAlike 4.0 International License. The
markdown+Rknitr source code of this blog is available under a GNU General Public
License (GPL v3) license from github.
Introduction
Statistics 101 teaches that for a distribution, possibly contaminated with outliers, a robust measure of the central tendency is the median. Not knowing this fact can make your analysis worthy to report in the newspaper (Google translate).
Higher quantiles of a distribution also have a long history as threshold for when to declare an observation an outlier. For example, growth curves for children illustrate how the quantiles of, e.g., the BMI distribution develop by age. Obesity is then for children defined as exceedance of the 97.7% quantile of the distribution at a particular age. Quantile regression is a non-parametric method to compute such curves and the statistical community has been quite busy lately investigating new ways to compute such quantile regressions models.
The focus of this blog post is nevertheless the simplest setting: Given an iid. sample \(\bm{x}\) of size \(n\) from a univariate and absolutely continuous distribution \(F\), how does one compute an estimate for the \(p\)-Quantile of \(F\) together with a corresponding two-sided \((1-\alpha)\cdot 100\%\) confidence interval for it?
The Point Estimate
Computing the quantile in a sample with statistical software is
discussed in the excellent survey of Hyndman and Fan (1996). The simplest
estimator is based on the order statistic
of the sample, i.e. \(x_{(1)} < x_{(2)}
< \cdots < x_{(n)}\). \[
\hat{x}_p = \min_{k} \left\{\hat{F}(x_{(k)}) \geq p\right\} = x_{(\lceil
n \cdot p\rceil)},
\] where \(\hat{F}\) is the empirical
cumulative distribution function of the sample. Since \(\hat{F}\) has jumps of size \(1/n\) the actual value of \(\hat{F}(\hat{x}_{p})\) can end up being
somewhat larger than the desired \(p\).
Therefore, Hyndman
and Fan (1996) prefer estimators interpolating between the two
values of the order statistic with \(\hat{F}\) just below and just above \(p\). It is interesting that even 20
years after, there still is no universally accepted way to do this
in different statistical software and the type argument of
the quantile function in R has been a close friend when
comparing results with SPSS or Stata users. In what follows we will,
however, stick with the simple \(x_{(\lceil n
\cdot p\rceil)}\) estimator stated above.
Below is illustrated how one would use R to compute the, say, the 80%
quantile of a sample using the above estimator. We can compute this
either manually or using the quantile function with
type=1:
##Make a tiny artificial dataset, say, the BMI z-score of 25 children
sort(x <- rnorm(25))## [1] -1.44090165 -1.40318433 -1.21953433 -0.95029549 -0.90398754 -0.66095890 -0.47801787
## [8] -0.43976149 -0.36174823 -0.34116984 -0.33047704 -0.31576897 -0.28904542 -0.03789851
## [15] -0.03764990 -0.03377687 0.22121130 0.30331291 0.43716773 0.47435054 0.60897987
## [22] 0.64611097 1.20086374 1.52483138 2.67862782##Define the quantile we want to consider
p <- 0.8
##Since we know the true distribution we can easily find the true quantile
(x_p <- qnorm(p))## [1] 0.8416212##Compute the estimates using the quantile function and manually
c(quantile=quantile(x, type=1, prob=p), manual=sort(x)[ceiling(length(x)*p)])## quantile.80% manual
## 0.4743505 0.4743505Confidence interval for the quantile
Besides the point estimate \(\hat{x}_p\) we also would like to report a
two-sided \((1-\alpha)\cdot 100\%\)
confidence interval \((x_p^{\text{l}},
x_p^{\text{u}})\) for the desired population quantile. The
interval \((x_p^{\text{l}},
x_p^{\text{u}})\) should, hence, fulfill the following condition:
\[
P( (x_p^{\text{l}}, x_p^{\text{u}}) \ni x_p) = 1 - \alpha,
\] where we have used the “backwards” \(\in\) to stress the fact that it’s
the interval which is random. Restricting the limits of this
confidence intervals to be one of the realisations from the
order statistics implies that we need to find indices \(d\) and \(e\) with \(d<e\) s.t. \[
P( x_{(d)} \leq x_p \leq x_{(e)}) \geq 1 - \alpha.
\] Note that it may not be possible to achieve the desired
coverage exactly in this case. For now we prefer the conservative choice
of having to attain at least the desired coverage. Note
that for \(1\leq r \leq n\) we have
\[
\begin{align*}
P( x_{(r)} \leq x_p) &= P(\text{at least $r$ observations are
smaller than or equal to $x_p$}) \\
&= \sum_{k=r}^{n} P(\text{exactly $k$ observations are smaller
than or equal to $x_p$}) \\
&= \sum_{k=r}^{n} {n \choose k} P(X \leq x_p)^k (1-P(X \leq
x_p))^{n-k} \\
&= \sum_{k=r}^{n} {n \choose k} p^k (1-p)^{n-k} \\
&= 1 - \sum_{k=0}^{r-1} {n \choose k} p^k (1-p)^{n-k}
\end{align*}
\] In principle, we could now try out all possible \((d,e)\) combinations and for each interval
investigate, whether it has the desired \(\geq
1-\alpha\) property. If several combinations achieve this
criterion we would, e.g., take the interval having minimal length. This
is what the MKmisc::quantileCI function does. However, the
number of pairs to investigate is of order \(O(n^2)\), which for large \(n\) quickly becomes lengthy to compute.
Instead, we compute an equi-tailed confidence interval
by finding two one-sided \(1-\alpha/2\)
intervals, i.e. we find \(d\) and \(e\) s.t. \(P(x_{(d)} \leq x_p) = 1-\alpha/2\) and
\(P(x_p \geq x_{(e)}) = 1-\alpha/2\).
In other words,
\[ \begin{align*} d &= \argmax P(x_{(r)} \leq x_p) \geq 1 - \frac{\alpha}{2} \\ &= \texttt{qbinom(alpha/2, size=n, prob=p)} \\ e &= \argmin P(x_p \geq x_{(r)}) \geq 1 - \frac{\alpha}{2} \\ &= \texttt{qbinom(1-alpha/2, size=n, prob=p) + 1} \end{align*} \]
Note that the problem can arise, that the above solutions are zero or
\(n+1\), respectively. In this case one
has to decide how to proceed. For an illustration of the above in case
of the median see the post by @freakonometrics. Also
note that the qbinom function uses the Cornish-Fisher
Expansion to come up with an initial guess for the quantile, which
is then refined by a numerical search. In other words, the function is
of order \(O(1)\) and will, hence, be
fast even for large \(n\).
When it comes to confidence intervals for quantiles the set of alternative implementations in R is extensive. Searching for this on CRAN, we found the following functionality:
| Package::Function | Version | Description |
|---|---|---|
MKmisc::quantileCI |
Implements an exact but very slow \(O(n^2)\) search as well as an asymptotic
method approximating the exact procedure. Due to the method being slow
it is not investigated further, but looking at it an Rcpp
implementation of the nested loop might be able to speed up the
performance substantially. Note: New versions of MKmisc do
not install, because the depency pkg limma is not on CRAN
anymore. |
|
jmuOutlier::quantileCI |
2.2 | Implements the exact method. |
envStats::eqnpar |
2.1.1 | implements both an exact and an asymptotic interval |
asht::quantileTest |
0.9.6 | also implements an exact method |
Qtools::confint.midquantile |
operates on the mid-quantile (whatever that is). The method is not investigated further. | |
There might even be more, but for now we are satisfied comparing just the above mentioned procedures:
as.numeric(jmuOutlier::quantileCI(x=x, probs=p, conf.level=0.95)[1,c("lower","upper")])
as.numeric(EnvStats::eqnpar(x=x, p=p, ci=TRUE, ci.method="exact",approx.conf.level=0.95)$interval$limits)
as.numeric(EnvStats::eqnpar(x=x, p=p, ci=TRUE, ci.method="normal.approx",approx.conf.level=0.95)$interval$limits)
as.numeric(asht::quantileTest(x=x,p=p,conf.level=0.95)$conf.int)## [1] -0.03377687 1.52483138
## [1] 0.2212113 2.6786278
## [1] -0.03377687 1.52483138
## [1] -0.03377687 2.67862782An impressive number of similar, but yet, different results! To add
to the confusion here is our take at this as developed in the
quantileCI package available from github:
devtools::install_github("hoehleatsu/quantileCI")The package provides three methods for computing confidence intervals for quantiles:
quantileCI::quantile_confint_nyblom(x=x, p=p, conf.level=0.95,interpolate=FALSE)
quantileCI::quantile_confint_nyblom(x=x, p=p, conf.level=0.95,interpolate=TRUE)
quantileCI::quantile_confint_boot(x, p=p, conf.level=0.95,R=999, type=1)## [1] -0.03377687 2.67862782
## [1] 0.07894831 1.54608644
## [1] -0.03377687 1.20086374The first procedure with interpolate=FALSE implements
the previously explained exact approach, which is also implemented in
some of the other packages. However, when the interpolate
argument is set to TRUE (the default), an additional
interpolation step between the two neighbouring order statistics is
performed as suggested in the work of Nyblom (1992), which extends work for the
median by Hettmansperger and Sheather
(1986) to arbitrary quantiles. It generates intervals of the
form:
\[ \left( (1-\lambda_1) x_{(d)} + \lambda_1 x_{(d+1)}, (1-\lambda_2) x_{(e-1)} + \lambda_1 x_{(e)} \right) \]
with \(0 \leq \lambda_1, \lambda_2 \leq 1\) chosen appropriately to get as close to the desired coverage as possible without knowing the exact underlying distribution - see the paper for details.
The last call in the above is to a basic bootstrap procedure, which
resamples the data with replacement, computes the quantile using
type=1 and then reports the 2.5% and 97.5% percentiles of
this bootstrapped distribution. Such percentiles of the basic bootstrap
are a popular way to get confidence intervals for the quantile, e.g.,
this is what we have used in Höhle and Höhle (2009) for
reporting the 95% quantile of the absolute difference in height at so
called check points in the assessment of accuracy for a
digital elevation model (DEM) in photogrammetry. However, the coverage
of the percentile bootstrap procedure is not without problems, because
the convergence rate as a function of the number of replicates \(r\) is only of order \(O(r^{-\frac{1}{2}})\) for quantiles (Falk and Kaufmann
(1991)). As a trade-off between accuracy and speed we use \(R=999\) throughout this post.
Simulation Study to Determine Coverage
We write a function, which for a given sample x computes
two-sided confidence intervals for the p-Quantile using a selection of
the above described procedures:
quantile_confints(x, p=p, conf.level=0.95)## jmuOutlier_exact EnvStats_exact EnvStats_asymp asht_quantTest nyblom_exact
## 1 -0.03377687 0.2212113 -0.03377687 -0.03377687 -0.03377687
## 2 1.52483138 2.6786278 1.52483138 2.67862782 2.67862782
## nyblom_interp boot
## 1 0.07894831 -0.03377687
## 2 1.54608644 1.49641655In order to evaluate the various methods and implementations we
conduct a Monte Carlo simulation study to assess each methods’ coverage.
For this purpose we write a small wrapper function to conduct the
simulation study using parallel
computation. The function wraps the
quantileCI::qci_coverage_one_sim function, which lets the
user define a simulation scenario (true underlying distribution, size of
the sample, etc.), then applies all confidence interval methods gathered
in the above quantile_confints and finally assesses whether
each confidence interval covers the true value or not.
simulate.coverage_qci <- function(n=n,p=p,conf.level=0.9, nSim=10e3, ...) {
##Windows users: change to below lapply function or use snow.
##lapplyFun <- function(x, mc.cores=NA, ...) pblapply(x, ...)
lapplyFun <- parallel::mclapply
sims <- dplyr::bind_rows(
lapplyFun(1L:nSim, function(i) {
quantileCI::qci_coverage_one_sim(qci_fun=quantile_confints, n=n,p=p,conf.level=conf.level,...)
}, mc.cores = parallel::detectCores() - 1)
) %>% summarise(across(everything(), mean))
return(sims)
}Simulation 1
We can now compare the coverage of the different implementation for
the particular n=25 and p=0.8 setting:
simulate.coverage_qci(n=25, p=0.8, conf.level=0.95)## jmuOutlier_exact EnvStats_exact EnvStats_asymp asht_quantTest nyblom_exact
## 1 0.9552 0.9461 0.9552 0.9786 0.9786
## nyblom_interp boot
## 1 0.9495 0.9198Note that the nyblom_interp procedure is closer to the
nominal coverage than it’s exact cousin nyblom_exact and
the worst results are obtained by the bootstrap percentile method. We
also note that the results of the jmuOutlier_exact method
appear to deviate from asht_quantTest as well as
nyblom_exact, which is surprising, because they should
implement the same approach.
Simulation 2
As a further test-case we consider the situation for the median in a smaller sample:
simulate.coverage_qci(n=11, p=0.5, conf.level=0.95)## jmuOutlier_exact EnvStats_exact EnvStats_asymp asht_quantTest nyblom_exact
## 1 0.9873 0.9332 0.9583 0.9873 0.9873
## nyblom_interp boot hs_interp
## 1 0.9463 0.9332 0.9463We note that the EnvStats_exact procedure again has a
lower coverage than the nominal required level, it must therefore
implement a slightly different procedure than expected. That coverage is
less than the nominal for an exact method is, however, still
somewhat surprising.
Edit 2019-05-14: I pointed the package maintainer to
this problem after looking at the code. Versions larger than 2.1.1 of
the EnvStats pkg now contain an updated version of this
function and also the Nyblom
function.
In this study for the median, the original Hettmansperger and Sheather
(1986) procedure implemented in quantileCI as
function median_confint_hs is also included in the
comparison (hs_interp). Note that the Nyblom (1992) procedure
for \(p=\frac{1}{2}\) just boils down
to this approach. Since the neighbouring order statistics are combined
using a weighted mean, the actual level is just close to the nominal
level. It can, as observed for the above setting, be slightly lower than
the nominal level. The bootstrap method again doesn’t look too
impressive.
Simulation 3
We finally also add one of the scenarios from Table 1 of the Nyblom (1992) paper, which allows us to check our implementation against the numerical integration performed in the paper to assess coverage.
simulate.coverage_qci(n=11, p=0.25, conf.level=0.90)## jmuOutlier_exact EnvStats_exact EnvStats_asymp asht_quantTest nyblom_exact
## 1 0.9219 0.7633 0.841 0.9219 0.9219
## nyblom_interp boot
## 1 0.9014 0.8816In particular the results of EnvStats_exact look
disturbing. The coverage of the interpolated order statistic approach
again looks convincing.
Simulation 4
Finally, a setup with a large sample, but now with the t-distribution with one degree of freedom:
simulate.coverage_qci(n=101, p=0.9, rfunc=rt, qfunc=qt, conf.level=0.95, df=1)## jmuOutlier_exact EnvStats_exact EnvStats_asymp asht_quantTest nyblom_exact
## 1 0.9547 0.9352 0.9547 0.9547 0.9547
## nyblom_interp boot
## 1 0.9495 0.9362Again the interpolation method provides the most convincing results. The bootstrap on the other hand again has the worst coverage, a larger \(R\) might have helped here, but would have made the simulation study even more time consuming.
Conclusion and Future Work
Can we based on the above recommend one procedure to use in practice?
Well, even though the simulation study is small, the exact
EnvStats::eqnpar approach appears to yield below nominal
coverage intervals, sometimes even substantially, and hence is not
recommended in versions below or equal to 2.1.1 1. On
the other hand, jmuOutlier_exact,
asht_quantTest, and nyblom_exact in all four
cases provide above nominal level coverage, i.e. the intervals are
conservative in as much as they are too wide. Also slightly disturbing
is that the results of the exact confidence interval methods varied
somewhat between the different R implementations. Part of the
differences arise from handling the discreteness of the procedure as
well as the edge cases differently. The basic percentile bootstrap
method is a simple approach, providing acceptable, but not optimal
coverage and also depends in part on \(R\). In particular for very large \(n\) or for a large number of replication in
the simulation study, the method with a large \(R\) can be slow. Suggestions exist in the
literature on how to improve the speed of coverage convergence by
smoothing (see, e.g., De Angelis, Hall, and Young
(1993)), but such work is beyond the scope of this post.
The Hettmansperger and Sheather
(1986) and Nyblom
(1992) method, respectively, provide very good coverage close to
the nominal level. The method is fast to compute, available through the
quantileCI R package and would be our recommendation to use
in practice.
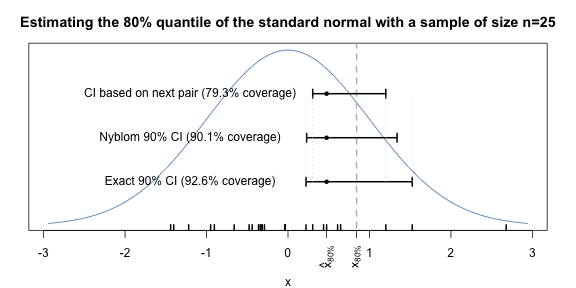
Altogether, we summarise our findings as follows: More confidence in confidence intervals for quantiles! and let the following picture illustrating 90% confidence intervals for the 80% quantile of the standard normal distribution based on the above sample of size \(n\)=25 say this in less than 1000 words.

Appendix
Given PDF \(f\) and CDF \(F\) of the underlying distribution, the coverage probability of the one sided Nyblom \((1-\alpha/2)\cdot 100\%\) confidence interval \((1-\lambda) x_{(r)} + \lambda x_{(r+1)}\) for \(x_p\) can be found as follows: Let \(z = (1-\lambda) x_{(r)} + \lambda x_{(r+1)}\). If the considered interval is a lower-confidence interval, we are interested in finding \(P( z \leq x_p)=\int_{-\infty}^{x_p} f_z(z) dz\). Here, the PDF of \(z\) is found as
\[ f_z(z) = \int_{-\infty}^{z} f_{x_{(r)},x_{(r+1)}}( x_{(r)}, x_{(r+1)}^*) dx_{(r)}, \quad\text{where}\quad x_{(r+1)}^* = \frac{z-(1-\lambda)x_{(r)}}{\lambda}. \]
The joint distribution of the order statistic is here
\[
f_{x_{(r)},x_{(r+1)}}( x_{(r)},
x_{(r+1)}) = \frac{n!}{(r-1)!(n-r-1)!} F(x_{(r)})^{r-1}
\left( 1- F(x_{(r+1)})\right)^{n-r-1} f(x_{(r+1)}) f(x_{(r)}).
\] The two nested integrals can be solved by numerical
integration using, e.g., the integral function.