Naming Uncertainty by the Bootstrap
Abstract
Data on the names of all newborn babies in Berlin 2016 are used to
illustrate how a scientific treatment of chance could enhance rank
statements in, e.g., onomastics investigations. For
this purpose, we first identify different stages of the naming-your-baby
process, which are influenced by chance. Second, we compute confidence
intervals for the ranks based on a bootstrap procedure reflecting the
above chance elements. This leads to an alternative league table based
on what we will call uncertainty corrected ranks. From
an R perspective we use the problem as a practice session for wrangling
data dplyr-style (code available by clicking on the github
logo in the license below).
 This work is licensed under a Creative Commons
Attribution-ShareAlike 4.0 International License. The
markdown+Rknitr source code of this blog is available under a GNU General Public
License (GPL v3) license from github.
This work is licensed under a Creative Commons
Attribution-ShareAlike 4.0 International License. The
markdown+Rknitr source code of this blog is available under a GNU General Public
License (GPL v3) license from github.
Introduction
What’s the most popular first name given to newborn boys and girls? This question seems to fascinate at different levels of temporal and spatial aggregation, because the choice of names and its dynamics reflects cultural and social behavior. The branch of science related to the study of names is entitled onomastics. Mathematical modelling is used in onomastics to study name dynamics by evolutionary models and models for contagious phenomena (Hahn and Bentley 2003; Kessler et al. 2012). But even the task of naming your baby has almost onomastics optimizing flavour requiring data science skills. However, once the Official Social Security Administration has released the numbers for all names of newborns in a given year, finding the most popular baby name appears a simple counting and ranking job: for example the most popular US baby names in 2015 were Emma (girls) and Noah (boys).

Statistics is the scientific study of chance. One fundamental concept is the inference of a population quantity from observing the quantity in a sample (=subset) of this population. To make this specific for the baby names: In Germany there is no official first name statistics, as a consequence, the site www.beliebte-vornamen.de uses information from a sample of 196,158 kids (corresponding to 26% of all newborns in Germany 2016) originating from a selection of registrar’s offices and birth clinics to determine the most popular first name in Germany 2016. However, the aspect of uncertainty in the resulting ranking, due to only measuring a sample of the population, is ignored when reporting the 2016 league table. The aspect of uncertainty can, however, also be more subtle. As an example, the city of Berlin recently released the official 2016 first name statistic of all newborns in the city (Landesamt für Bürger- und Ordnungsangelegenheiten Berlin 2017). The data are available at district level, which is helpful, because there are notable socio-economic and cultural differences between the districts. One could argue that since the data cover the entire population of interest (i.e. newborns in Berlin 2016) the use of inferential statistics is superfluous. But is it that simple?
In what follows we use the Berlin newborn names to illustrate how a scientific treatment of chance could enhance rank statements in general and in name rank tables in particular.
Descriptive Data Analysis
Altogether, the distrNames variable contains the
information about the frequency of 13245 unique first names. Below is
shown the first 10 lines of the data.

By summing the count column it becomes clear that in
total, 69525 names were registered in Berlin 2016 (35620 boys and 33905)
girls. The proportion of boy names is 51.2%. One caveat with the Berlin
name statistic is that, if a child is given several first names, each
name is counted once in the statistic. Hence, the above total sum is
actually higher than the number of kids born 2016 (38,030 in 2015,
official 2016 number not available yet). Despite of the potential
problems with multiple names per kids, the empirical boy fraction in the
data is close to reported ratios of the number of born boys vs. girls of
1.05 (Jacobsen, Møller,
and Mouritsen 1999), which means that the expected fraction of
boys among the newborns should be approximately 51.2%.
Strangely enough, 15 babies seem to have an empty first name (but the
sex is known). We decided to keep these NA names in the
analysis, because at the time of writing it was unclear, if this is a
data recording problem (e.g. a delay of the December 2016 kids) or
actually allowed. An email inquiry with the data providing agency
revealed that an empty name is the result of the naming authority declining
a chosen first name in the interest of the kid. In this case the
baby remains nameless on the birth certificate until the dispute is
resolved before court.
We can now look at the top-5-names in Berlin for each gender:
##Aggregate data over district and sort according to rank within gender
newborn <- distrNames %>% group_by(firstname, sex) %>%
summarise(count=sum(count)) %>%
arrange(desc(count)) %>% group_by(sex) %>%
mutate(rank=rank(-count,ties="min"))## Source: local data frame [10 x 4]
## Groups: sex [2]
##
## firstname sex count rank
## <chr> <fctr> <int> <int>
## 1 Marie f 695 1
## 2 Sophie f 649 2
## 3 Charlotte f 495 3
## 4 Maria f 403 4
## 5 Emilia f 382 5
## 6 Alexander m 467 1
## 7 Paul m 383 2
## 8 Elias m 371 3
## 9 Maximilian m 344 4
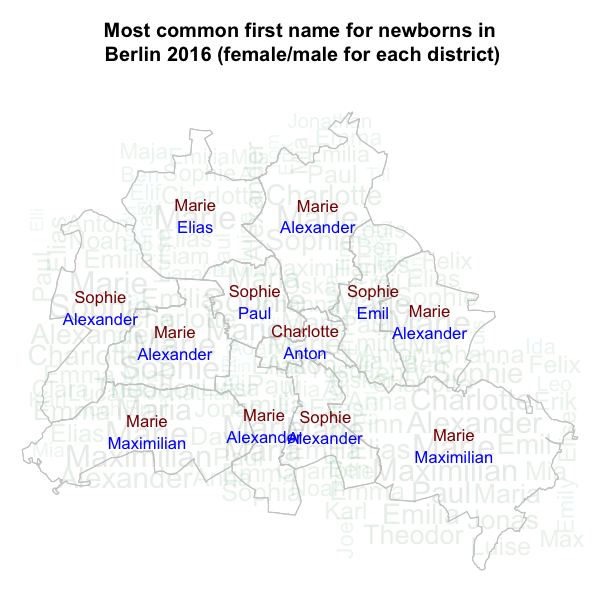
## 10 Emil m 295 5The top-1 names per gender and district from distrNames
can easily be computed in similar fashion using group_by
and summarise operations. To spice up the visualization we
use a custom made wordmapcloud, which overlays the
top-1 names over an alpha-channeled wordcloud of the district’s name
with font size proportional to frequency. In the resulting plot we see
little geographical variation in the top-1 names over the districts -
particularly for girls.

The Gini
index for the name frequency is calculated using the
ineq package and is 0.728 and 0.743 for girls and boys,
respectively. This means that the occurrence of names in boys is
dominated by fewer names for boys than for girls. Furthermore, both
gender’s name distribution tend to be dominated by few names. This
feature can also be visualized by a Lorenz curve - here shown separately
for each sex:

From the curve one can for example deduce that the frequency of the top-50 girl names (top 0.7% out of the 6957 girl names), cover 29.0% of all 33905 girl namings.
Analysing Stochasticity in the Name Selection
At which places is stochasticity a useful concept for abstracting unobservable factors influencing the name selection? We shall focus on 5 stages:
the number of babies born in Berlin in 2016
the gender of a given born baby; as mentioned above the odds for the kid being a boy is roughly 1.05:1.
the number of names given to a baby of a specific sex
the selection of the name(s) given that the gender of the baby is known
reporting problems leading to the wrong name(s) being recorded
We will ignore uncertainty from stages 1, 3 and 5 and, hence, only focus on uncertainty arising from stages 2 and 4. One may ask in stage 4, if the naming is not deterministic, once the parents know the sex of their baby? In this post we take the position that even given sex the naming is the outcome of a stochastic process. The selection probabilities are likely to vary from couple to couple based on complex interactions between, e.g., social status, existing names in the family as well as past exposure and associations with names. Since data are never going to be available on these individual factors, we will, as a proxy, assume that the drawing probabilities are district specific. As a result, the selected name can be considered as one realization of the multinomial distribution with the underlying true popularity of all possible names in the district acting as selection probabilities.
Uncertainty Assessment using the Bootstrap
When combining the above stages 3 and 4, the name selection process
can be mimicked by a simple bootstrap procedure
stratified by district (Davison and Hinkley 1997). In
spirit, this approach corresponds to the bootstrap approach to ranks
used in Sect. 5.3 of Goldstein and Spiegelhalter
(1996). We operationalise this in R using the boot
package, the work-horse will be the function name_ranks
shown below.
######################################################################
## Compute rank of name within female and male population,
## respectively for a draw of all kids (one kid per row) with
## replacement.
##
## Parameters:
## x - the full data, one row per kid
## idx - vector of length nrow(x) containing a possible permutation
## (with replacement)
## returns - which column to return, "rank" or "count" (for use in boot).
## If returns=="dplyr::everything()", then entire frame is returned (useful for
## use with broom)
##
## Returns:
## vector or data.frame with stratified ranks (arranged by (firstname, sex))
######################################################################
name_ranks <- function(x, idx=seq_len(nrow(x)), returns=c("rank","count","dplyr::everything()")) {
##Make resampled data and append all_strata to ensure each firstname-sex combination occurs
x_boot <- x %>% slice(idx) %>% bind_rows(all_strata)
##Summarise the number of occurrences for each firstname-sex strata and compute the ranks.
aggrx_wranks <- x_boot %>% group_by(firstname,sex) %>%
summarise(count = sum(count)) %>%
group_by(sex) %>%
mutate(rank=rank(-count, ties.method="min")) %>%
arrange(firstname, sex) #important to ensure order.
##Select relevant columns
res <- aggrx_wranks %>% ungroup() %>% select_(returns)
##Return as vector (needed for boot pkg) or data.frame (needed from broom)
if (returns[1] %in% c("rank","count")) return(res %>% .[[1]]) else return(res)
}In the above, all_strata is a data.frame
containing all possible strata of gender and firstname. This is done in
order to ensure that we later get a zero count for names, even if they
do not appear in the bootstrap re-sample.
We then convert the aggregated data to long format where each kid is represented by one row. This is the most didactic way to explain what is going on in the bootstrap, but an aggregated multinomial approach would probably be faster in terms of execution time.
kids <- distrNames %>% slice(rep(seq_len(nrow(distrNames)), times=distrNames %$% count)) %>% mutate(count=1)Ready to perform the bootstrap stratified within districts? Yes, its
conveniently done using the boot package (which is easily
paralleled too).
set.seed(123) ##fix seed for reproducibility
b <- boot::boot(kids, statistic=name_ranks, R=999, strata=kids$district, parallel="multicore",ncpus=3)We use the percentile method on the 999 + 1 bootstrap rank-vectors as a method for computing a 90% confidence interval for the rank of each name for boys and girls, respectively.
Update 2017-02-07: Maëlle made me aware
of some newer ways to perform the bootstrap, e.g., using the
broom package. It’s especially useful for the parametric
bootstrap, but by joining with the previously calculated observed ranks,
the code for making a simple bootstrap stratified bootstrap actually
looks quite nice (although not parallized and hence slower):
require(broom)
b_broom <- kids %>% group_by(district) %>% bootstrap(m=999, by_group=TRUE) %>%
do({ name_ranks(.,returns="dplyr::everything()") }) %>%
group_by(sex,firstname) %>%
summarise("rankci.5."=quantile(rank, 0.05,type=3),"rankci.95."=quantile(rank, 0.95,type=3))
newborn_ranks <- newborn %>% inner_join(b_broom,by=c("firstname","sex")) %>% arrange(rank,sex)## Source: local data frame [10 x 6]
## Groups: sex [2]
##
## firstname sex count rank rankci.5. rankci.95.
## <chr> <fctr> <int> <int> <int> <int>
## 1 Marie f 695 1 1 2
## 2 Alexander m 467 1 1 1
## 3 Sophie f 649 2 1 2
## 4 Paul m 383 2 2 3
## 5 Charlotte f 495 3 3 3
## 6 Elias m 371 3 2 4
## 7 Maria f 403 4 4 5
## 8 Maximilian m 344 4 3 4
## 9 Emilia f 382 5 4 5
## 10 Emil m 295 5 5 9Using the lower limit of the 90% CI to group the names, we define the concept of a uncertainty corrected rank (ucrank). This is just the lowest rank which we, given the modelled stochasticity, cannot be ruled out (at the 5% lvl. of significance). Listing the top-5 of these corrected ranks leads to the following tables for girls and boys, respectively:
| ucrank (among girls) | first names (girls) | ucrank (among boys) | first names (boys) |
|---|---|---|---|
| 1 | Marie, Sophie | 1 | Alexander |
| 3 | Charlotte | 2 | Paul, Elias |
| 4 | Maria, Emilia | 3 | Maximilian |
| 6 | Anna, Emma, Mia, Sophia | 5 | Emil, Noah, Anton, Felix |
| 8 | Johanna, Luise | 6 | Oskar |
Instead of using the uncertainty corrected ranks, we could instead have visualized the 90% rank confidence intervals instead (dots denote the observed ranks):

Discussion
In this post we have used the bootstrap method as a way to assess uncertainty in ranks. This approach is very general and can be extended to areas beyond onomastics. No matter the area of application the approach requires a careful identification of the elements of chance you want to take into account. In the particular application we decided to ignore specific uncertainty aspects (e.g. number of babies born) to not impose further hard-to-verify assumptions. However, as soon as there is uncertainty, ranks are known to be subject to large variation. Hence, a different reporting or visualization of the ranks than the point estimator from the sample is necessary. The use of uncertainty corrected ranks is not revolutionary, but it underlines the importance of uncertainty in the construction of league tables. A more uncertainty enhancing presentation of ranks in, e.g., data journalism, is therefore needed.
